
In this simple exercise, students simulate how restriction enzymes and gel electrophoresis can be used to identify different species of penguins.
The first section describes how DNA fingerprints are made using restriction enzymes and gel electrophoresis.
A technique called polymerase chain reaction (PCR) is used to amplify the sequences of DNA.
Analysis also involves treating DNA with special enzymes called restriction enzymes, which act like molecular scissors to cut DNA at specific sites. Digestion by these enzymes will produce fragments of varying lengths.
If a restriction site occurs within the sample sequence, the probe will reveal multiple bands of DNA that have
different sizes. This technique is called restriction fragment length polymorphism (RFLP).
Students label an image that outlines the steps in the process.
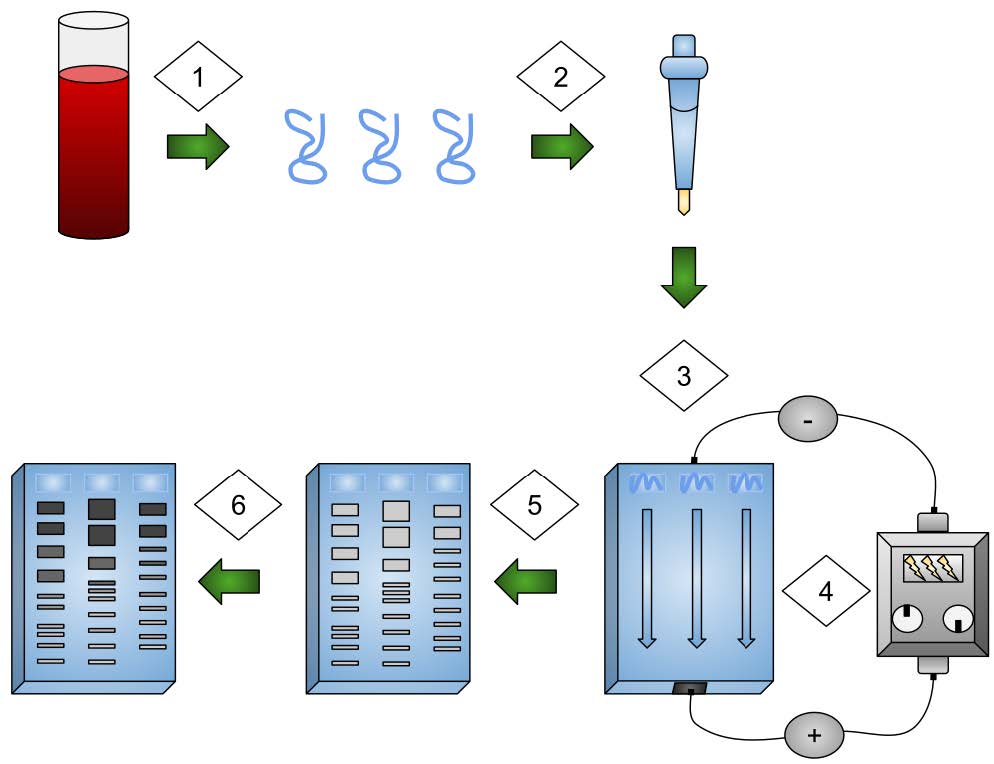
Using Fragments to Identify Penguins
The next section describes how the HaeIII enzyme cuts DNA at specific cites (GG/CC). Five sequences from different penguins are listed. Students find the restriction enzyme site in each sequence. This creates fragments of different lengths, or base-pairs.
Five species of penguins must be identified from the fragment lengths: Humboldt, Galapagos, Adelie, Emperor, and Little Blue.
By comparing the lengths of the fragments to the five penguins, students can identify which DNA sample belongs to which animal. This is a great way to introduce students to DNA biotechnology! Plus, who doesn’t love penguins!


